A posteriori estimator distinguishing different error components and adaptive stopping criteria
The $p$-Laplacian problem: distinguishing the algebraic, discretization, and linearization error components
Here, we will show the implementation work about the error estimator for the $p$-Laplacian problem which can distinguish the different error component in the computation, respectively the algebraic, linearization, and discretization error parts.
In the following, only some extractions of the full script will be shown to help understand the implementation work. You can download the full script at the end of this page.
We define the mesh, as for the $p$-Laplacian problem, we chose $p=10$ for example, so $q$ is given by $q=p/(p-1)$.
// definition of mesh int numnode=30; mesh Th=square(numnode,numnode); plot(Th,cmm="mesh",ps="mesh.eps"); // for the P-laplacian problem, we have 1/p+1/q=1 // the value for p and q. p/2 and q/2 //real numberp=10; // value of p real numberp=9; real numberpdiv2=numberp/2; // value of p/2 real numberq=numberp/(numberp-1); // value of q real numberqdiv2=numberq/2; // value of q/2
We use an analytic solution to test the performance of the adaptive inexact Newton method
// Data for given problem: exact solution // case for p!=2 func uExact = 1.0/numberq* ((0.5)^numberq-sqrt((x-0.5)^2+(y-0.5)^2)^numberq); // in the normal case func dxuExact = -0.5*((x-0.5)^2+(y-0.5)^2)^(numberqdiv2-1)*(2*x-1); func dyuExact = -0.5*((x-0.5)^2+(y-0.5)^2)^(numberqdiv2-1)*(2*y-1); func f=2;
We begin with an initial solution which is not far away from the exact solution, to do this, we add some oscillations by the parameters mu and lamda:
// to make an initial solution with some oscillation: lamda et mu real lamda = 1, mu = 0.5;
The classic Newton method to solve the $p$-laplacian problem is given by:
uhk = uExact*(1+4*lamda*x*(x-1)*y*(y-1)); // initial soluton which is close to the exact solution
func int SolverExactLinearCG()
{
varf VarfLaplacien(uh,vh)= int2d(Th)(((numberp-1)*(sqrt(Grad(uhk)'*Grad(uhk)))^(numberp-2))*Grad(uh)'*Grad(vh))
+int2d(Th)(f*vh+((numberp-2)*(sqrt(Grad(uhk)'*Grad(uhk)))^(numberp-2))*Grad(uhk)'*Grad(vh))
+on(1,2,3,4,uh=uExact);
varf VarfBC1(uh,vh)=on(1,2,3,4,uh=1);
matrix A=VarfLaplacien(Uh,Uh,tgv=-1,solver=GMRES);
real[int] b=VarfLaplacien(0,Uh,tgv=-1);
real[int] BC1=VarfBC1(0,Uh,tgv=-1);
real epsgc=1e-10;
func real[int] DJ(real[int] &u)
{
real[int] Au=A*u;
return Au;
}
int NumLinearization=0;
real eps1=1e-8;
for (int k=1; k<=1000; k++)
{
NumLinearization++;
A=VarfLaplacien(Uh,Uh,tgv=-1,solver=GMRES);
b=VarfLaplacien(0,Uh,tgv=-1);
uh[] = BC1 ? b : uhk[];
LinearCG(DJ,uh[],b,veps=epsgc,nbiter=1000,verbosity=nbverbosity);
epsgc=-abs(epsgc);
Uh udiff=uh-uhk;
if ((abs(udiff[].max) < eps1)||(abs(numberp-2) < eps1)) break;
uhk=uh;
}
udisc=uh;
}
SolverExactLinearCG();
We denote the different types of solution by:
Uh uh; // obtained solution at the (precedent+nu) step Uh uhk; // obtained solution at the LAST Newton linearization step by adaptive inexact method Uh udisc; // obtained solution by a exact algebraic solver and exact Newton linearization Uh uhinfiny; // obtained solution by a exact algebraic solver at each Newton linearization Uh uhp; // obtained solution at the precedent step
The functions to compute different types of error
// Computation of Total error: uExact -- uhp func real ComputeErrorTotal(int setValuePlot) // Computation of Discretization error: uExact -- uDisc func real ComputeErrorDisc(int setValuePlot) // Computation of Algebraic error: uDisc - uhinfiny func real ComputeErrorLin(int setValuePlot) // Computation of Linearization error: uhinfiny --uhp func real ComputeErrorAlg(int setValuePlot)
Adaptive inexact Newton method to solve the $p$-Laplacian problem
uhk = uExact*(1+4*lamda*x*(x-1)*y*(y-1)); // initiale soluton which is close to the exact solution
func int SolverInexactAdaptiveLinearCG()
{
varf VarfLaplacien(uh,vh)= int2d(Th)(((numberp-1)*(sqrt(Grad(uhk)'*Grad(uhk)))^(numberp-2))*Grad(uh)'*Grad(vh))
+int2d(Th)(f*vh) + int2d(Th)(((numberp-2)*(sqrt(Grad(uhk)'*Grad(uhk)))^(numberp-2))*Grad(uhk)'*Grad(vh))
+on(1,2,3,4,uh=uExact);
varf VarfBC1(uh,vh)=on(1,2,3,4,uh=1);
matrix A=VarfLaplacien(Uh,Uh,tgv=-1,solver=GMRES);
set(A,solver=CG);
real[int] b=VarfLaplacien(0,Uh,tgv=-1);
real[int] BC1=VarfBC1(0,Uh,tgv=-1);
func real[int] DJ(real[int] &u)
{
real[int] Au=A*u;
return Au;
}
// main part for the solving
NumLinearization=0;
real epsgc=1e-10;
for (int k=1; k<=1000; k++)
{
NumLinearization++;
A=VarfLaplacien(Uh,Uh,tgv=-1,solver=GMRES);
b=VarfLaplacien(0,Uh,tgv=-1);
uh[] = BC1 ? b : uhk[];
LinearCG(DJ,uh[],b,eps=eps1,nbiter=1000,verbosity=nbverbosity,stop=stop);
if(CritereArret == 3) break;
uhk=uh;
}
}
SolverInexactAdaptiveLinearCG();
The adaptive stopping criteria is defined in the "stop" function, which is given by:
func bool stop(int iter, real[int] u, real[int] g)
{
CritereArret = 10;
if (iter%nu==0 && iter > 1){
sigma1=(sqrt(Grad(uh)'*Grad(uh)))^(numberp-2)*dx(uh);
sigma2=(sqrt(Grad(uh)'*Grad(uh)))^(numberp-2)*dy(uh);
sigmak1=(numberp-1)*(sqrt( Grad(uhk)'*Grad(uhk)))^(numberp-2)*dx(uh)-(numberp-2)*(sqrt(Grad(uhk)'*Grad(uhk)))^(numberp-2)*dx(uhk);
sigmak2=(numberp-1)*(sqrt( Grad(uhk)'*Grad(uhk)))^(numberp-2)*dy(uh)-(numberp-2)*(sqrt(Grad(uhk)'*Grad(uhk)))^(numberp-2)*dy(uhk);
CritereArret=ComputeEstimator(iter);
if (CritereArret == 2 || CritereArret == 3) return 1;
}
return 0;
}
The function ComputeEstimator is the main part, which correspond the adaptive inexact Newton algorithm and the stopping criterion.
func int ComputeEstimator(int numiterCG)
{
if (numiterCG==nu){
RHSdplusl(); // compute d+l
RHSd(); // compute d
uhp=uh; // in each Newton linearization, we begin from uh
[d1p,d2p]=[d1,d2];
[dplusl1p,dplusl2p]=[dplusl1,dplusl2];
real etaDisc=ComputeEtaDisc(0);
real etaLin=ComputeEtaLin(0);
etaDiscSave=etaDisc;
etaLinSave=etaLin;
}else{
RHSdplusl(); //compute d+l
RHSd(); //compute d
real etaRem=ComputeEtaRem(0);
real etaAlg=ComputeEtaAlg(0);
real maxError1=max(etaDiscSave,etaLinSave);
real maxError2=max(maxError1,etaAlg);
if (etaRem>GammaRem*maxError2)
{
return 1;
}else{
if (etaAlg>GammaAlg*maxError1){
uhp=uh;
[d1p,d2p]=[d1,d2];
[dplusl1p,dplusl2p]=[dplusl1,dplusl2];
real etaDisc=ComputeEtaDisc(0);
real etaLin=ComputeEtaLin(0);
etaDiscSave=etaDisc;
etaLinSave=etaLin;
return 1;
}else{
if (etaLinSave<=GammaLin*etaDiscSave){
return 3;
}
uhk=uhp;
return 2;
}
}
}
}
We have four functions to compute the different estimators, respectively:
func real ComputeEtaDisc(int setValuePlot)
{
real etaDisc;
if (setValuePlot)
{
// varf indicatoretaDisc(unused,chiK) = int2d(Th)(chiK*(((sqrt(Grad(uh)'*Grad(uh))^(numberp-2)*Grad(uh)+[d1,d2])'* (sqrt(Grad(uh)'*Grad(uh))^(numberp-2)*Grad(uh)+[d1,d2]))^numberqdiv2));
varf indicatoretaDisc(unused,chiK) = int2d(Th)(chiK*(((sqrt(Grad(uhp)'*Grad(uhp))^(numberp-2)*Grad(uhp)+[d1,d2])'* (sqrt(Grad(uhp)'*Grad(uhp))^(numberp-2)*Grad(uhp)+[d1,d2]))^numberqdiv2));
Wh mapetaDisc;
mapetaDisc[]=indicatoretaDisc(0,Wh);
etaDisc=2^(1.0/numberp)*(mapetaDisc[].sum)^(1.0/numberq);
mapetaDisc=2^(1.0/numberp)*(mapetaDisc)^(1.0/numberq);
plot(mapetaDisc, ps="MapDiscEstimator.eps",value=1, fill=1,cmm="DiscretizationEstimator");
}else
{
etaDisc=int2d(Th)(((sqrt(Grad(uhp)'*Grad(uhp))^(numberp-2)*Grad(uhp)+[d1p,d2p])'* (sqrt(Grad(uhp)'*Grad(uhp))^(numberp-2)*Grad(uhp)+[d1p,d2p]))^numberqdiv2);
etaDisc=2^(1.0/numberp)*etaDisc^(1.0/numberq);
}
return etaDisc;
}
func real ComputeEtaAlg(int setValuePlot)
{
real etaAlg;
if (setValuePlot)
{
varf indicatoretaAlg(unused,chiK) = int2d(Th)(chiK*( ( ([dplusl1,dplusl2]-[dplusl1p,dplusl2p])'*([dplusl1,dplusl2]-[dplusl1p,dplusl2p]) )^numberqdiv2 ));
Wh mapetaAlg;
mapetaAlg[]=indicatoretaAlg(0,Wh);
etaAlg=(mapetaAlg[].sum)^(1.0/numberq);
mapetaAlg=(mapetaAlg)^(1.0/numberq);
plot(mapetaAlg, ps="MapAlgebraicEstimator.eps",value=1, fill=1,cmm="AlgebraicEstimator");
}else
{
etaAlg=int2d(Th)( ( ([dplusl1,dplusl2]-[dplusl1p,dplusl2p])'*([dplusl1,dplusl2]-[dplusl1p,dplusl2p]) )^numberqdiv2 );
etaAlg=etaAlg^(1.0/numberq);
}
return etaAlg;
}
func real ComputeEtaLin(int setValuePlot)
{
real etaLin;
if (setValuePlot)
{
varf indicatoretaLin(unused,chiK) = int2d(Th)(chiK*(( ([dplusl1p,dplusl2p]-[d1p,d2p])'*([dplusl1p,dplusl2p]-[d1p,d2p]))^numberqdiv2));
Wh mapetaLin;
mapetaLin[]=indicatoretaLin(0,Wh);
etaLin=(mapetaLin[].sum)^(1.0/numberq);
mapetaLin=(mapetaLin)^(1.0/numberq);
plot(mapetaLin, ps="MapLinearizationEstimator.eps",value=1, fill=1,cmm="LinearizationEstimator");
}else
{
etaLin=int2d(Th)(( ([dplusl1p,dplusl2p]-[d1p,d2p])'*([dplusl1p,dplusl2p]-[d1p,d2p]))^numberqdiv2 );
etaLin=etaLin^(1.0/numberq);
}
return etaLin;
}
func real ComputeEtaRem(int setValuePlot)
{
real[int] rh(Th.nt);
real[int] rhK(Th.nt); //vector rh multiply area of their triangle
rh = 0.;
for(int i=0; i < Th.nt; i++)
{
for(int j=0; j < 3; j++)
{
int indv = Th[i][j];
rh[i] += (Ra[indv]/Tha[indv].area);
}
rhK[i] = rh[i]*(Th[i].area)^(2.0/numberq);
}
real etaRem = (diameterArea * sqrt(rh'*rhK));
return etaRem;
}
Now, the point is how to construct the equilibrated flux, as denoted by $d+l$ and $d$ at $i$th step in the algebraic solver, as well as at $(i+nu)$-th step. In my script, I denoted them by
fespace Vh(Th,RT1); // Raviart-Thomas space // valeur de d et d_precedent Vh [d1,d2] = [0,0]; Vh [d1p,d2p] = [0,0]; // valeur de d+l et d+l_precedent Vh [dplusl1,dplusl2] = [0,0]; Vh [dplusl1p,dplusl2p] = [0,0];
Firstly, we construct the "patches" associated to each node in the mesh.
fespace Pa(ThaI,P1dc);
for(int i=0; i < Th.nv; ++i)
{
phia[][i]=1; // phia
Tha[i] = trunc (Th, phia > 0,label=10);
if (Th(i).label==0) Tha[i]=change(Tha[i],flabel=10);
phia[][i]=0; // phia
}
For the local problems, we have the same matrix during the algorithm, so we prepare it in advance
real epsregua = numnode*numnode*1e-8; //regularisation term to ensure the uniqueness of the solution of the local problem
matrix[int] Aa(Th.nv);
func int globalMatrix(matrix[int] & Aa)
{
for( int i=0; i < Th.nv ;++i)
{
phia[][i]=1; // phia
ThaI = Tha[i];
VPa [dIa1,dIa2,qa],[v1,v2,phih];
varf Varflocala([dIa1,dIa2,qa],[v1,v2,phih]) =
int2d(ThaI)( [dIa1,dIa2]'*[v1,v2] - qa*Div(v1,v2) - Div(dIa1,dIa2)*phih - epsregua*qa*phih )
+ on(10, dIa1=0,dIa2=0);
matrix Art=Varflocala(VPa,VPa);
Aa[i]= Art;
set(Aa[i],solver=sparsesolver);
phia[][i]=0;
}
}
globalMatrix(Aa);
Eachtime we do the estimation, we need to solve the two local problems, respectively for $d$ and $d+l$, as the matrix is same during the inexact Newton algorithm, only the right-hand-side change, we here call the functions RHSd() and RHSdplusl().
Attention, here we can do some optimizations for the code, as shown for the laplace problem. i.e. we can prepare a table of matrix associated to the RHS in advance, as well as the computational part for each estimator.
func int RHSd()
{
Vh [d11,d22]=[0,0];
int indv=0;
// boucle pour calculer le residu Rabas dans le probleme local d
for(int i=0; i < Th.nv; i++) {
phia[][i]=1;
ThaI = Tha[i];
Rabas[i] = int2d(ThaI)(f'*phia-(sqrt(Grad(uh)'*Grad(uh)))^(numberp-2)*(Grad(uh)'*Grad(phia)));
phia[][i]=0;
}
//set value of Rabas to zero for vertices lie on the boundary
indv = 0;
for(int i=0; i < Th.nbe; i++) {
for(int j=0; j < 2; j++) {
indv = Th.be(i)[j];
Rabas[indv] = 0.;
}
}
for(int i=0; i < Th.nv ;++i)
{
phia[][i]=1;
ThaI = Tha[i];
Pa Chia =1;
VPa [d1tmp,d2tmp,qa],[v1,v2,phih];
varf l([uv1,uv2,qa],[v1,v2,phih]) = -int2d(ThaI)( phia*(([sigma1,sigma2])'*[v1,v2] + f*phih )
-Grad(phia)'*[sigma1,sigma2]*phih - Rabas[i]*phih/ThaI.area)
+ on(10,uv1 = 0, uv2 = 0);
VPa [F,F1,F2] ;
F[] = l(0,VPa);
Fa[i].resize(F[].n);
Fa[i]=F[];
d1tmp[] = Aa[i]^-1*Fa[i];
real[int] so(d1tmp[].n);
so=d11[](I[i]);
d1tmp[]= d1tmp[].*epsI[i];
d1tmp[] += so;
d11[](I[i]) = d1tmp[];
phia[][i]=0;
}
[d1, d2]=[d11,d22];
return 1;
}
func int RHSdplusl()
{
Vh [dplusl11,dplusl22]=[0,0];
int indv=0;
//for loop computing vector Ra
for(int i=0; i < Th.nv; i++) {
phia[][i]=1;
ThaI = Tha[i];
Ra[i] = int2d(ThaI)(f'*phia
-(numberp-1)* (sqrt( Grad(uhk)'*Grad(uhk)))^(numberp-2)*(Grad(uh)'*Grad(phia))
+ (numberp-2)*(sqrt(Grad(uhk)'*Grad(uhk)))^(numberp-2)*(Grad(uhk)'*Grad(phia)));
phia[][i]=0;
}
//set value of Ra to zero for vertices lie on the boundary
indv = 0;
for(int i=0; i < Th.nbe; i++)
{
for(int j=0; j < 2; j++)
{
indv = Th.be(i)[j];
Ra[indv] = 0.;
}
}
for(int i=0; i < Th.nv ;++i)
{
phia[][i]=1;
ThaI = Tha[i];
Pa Chia =1;
VPa [dplusl1tmp,dplusl2tmp,qa],[v1,v2,phih];
varf l([uv1,uv2,qa],[v1,v2,phih]) = -int2d(ThaI)( phia*(([sigmak1,sigmak2])' *[v1,v2] + f*phih)
-Grad(phia)'*[sigmak1,sigmak2]*phih - Ra[i]*phih/ThaI.area)
+ on(10,uv1 = 0, uv2 = 0);
VPa [F,F1,F2] ;
F[] = l(0,VPa);
Fa[i].resize(F[].n);
Fa[i]=F[];
dplusl1tmp[] = Aa[i]^-1*Fa[i];
real[int] so(dplusl1tmp[].n);
so=dplusl11[](I[i]);
dplusl1tmp[]= dplusl1tmp[].*epsI[i];
dplusl1tmp[] += so;
dplusl11[](I[i]) = dplusl1tmp[];
phia[][i]=0;
}
[dplusl1,dplusl2]=[dplusl11,dplusl22];
return 1;
}
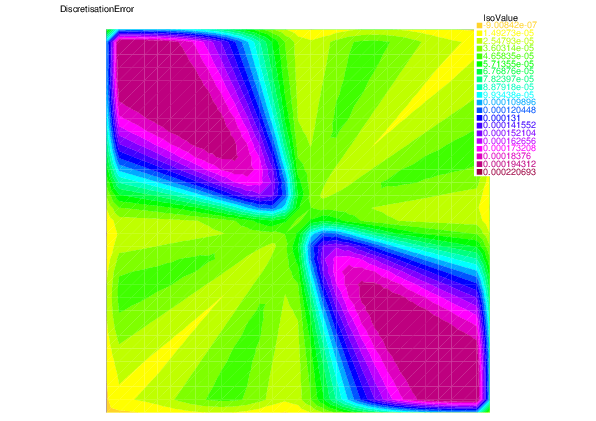
We see that for each error component: discretization, algebraic, and linearization errors, the overall error distributions are very well predicted by the corresponding estimators.
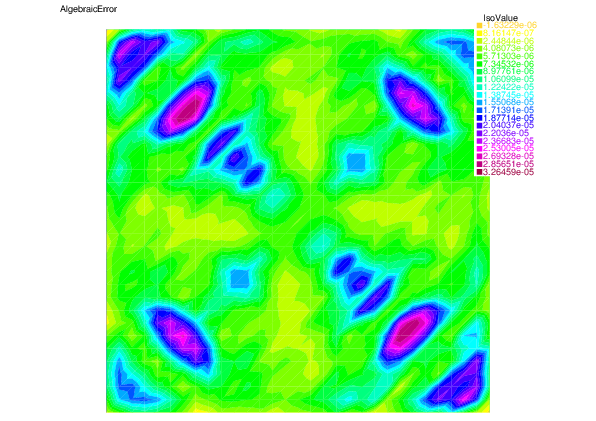 |
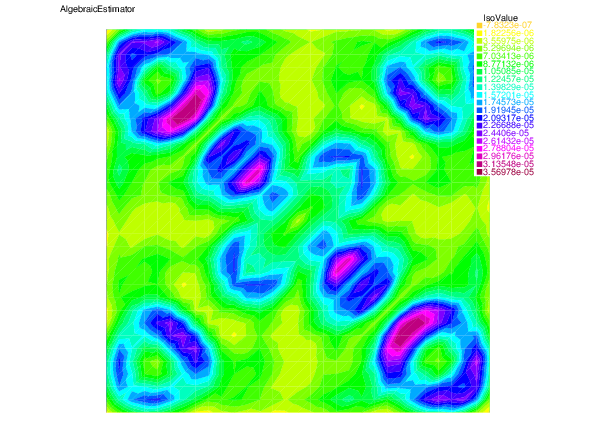 | |
| Algebraic error | Algebraic estimator |
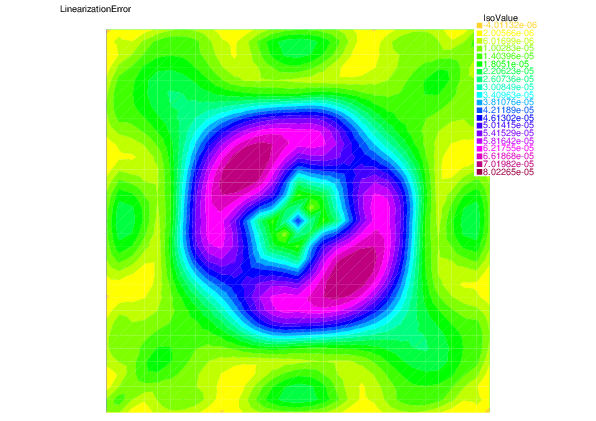 |
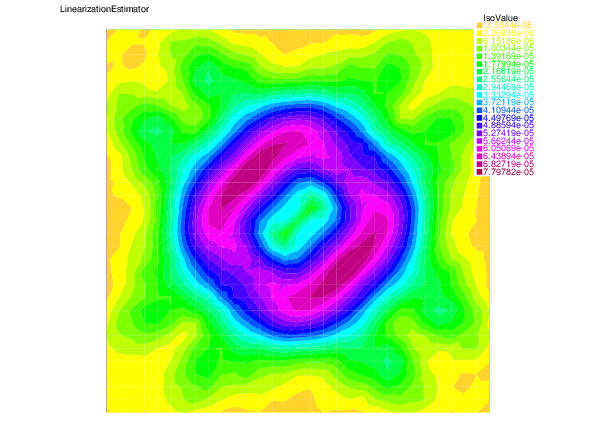 | |
| Linearization error | Linearization estimator |
 |
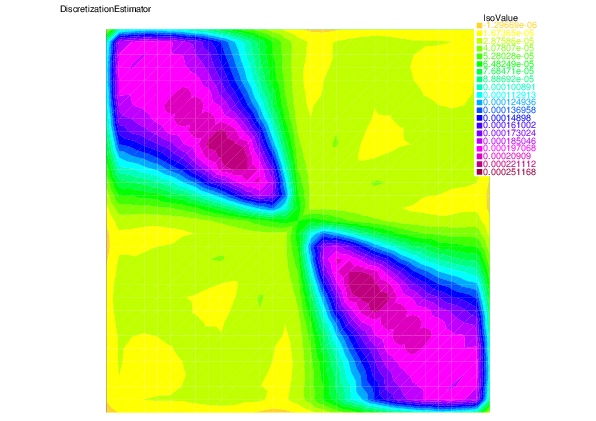 | |
| Discretization error | Discretization estimator |
download the script: ASCpLapalceI.edp or return to main page